Human Models
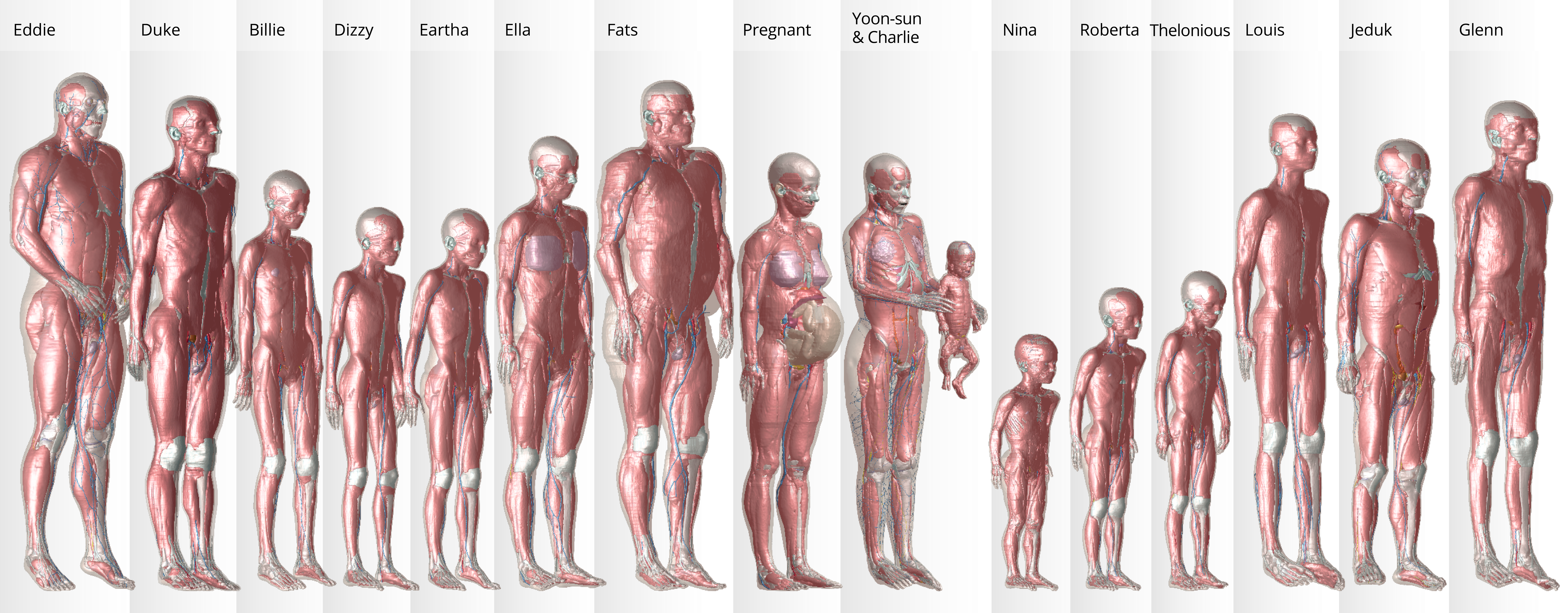
The Virtual Population (ViP) models are a set of detailed high-resolution anatomical models created from magnetic resonance image data of volunteers. Since their inception, the ViP models have become the gold standard for in silico biophysical modeling applications.
The newest generation of our phantoms is the ViP V3.x & V4.x. The V3.1 models elevate computational simulations in 3D anatomies to an unprecedented level of detail and accuracy with more than 300 tissues and organs per model and a resolution of 0.5 × 0.5 × 0.5 mm³ throughout the entire body. The V4.x models include detailed subdivisions of muscles, blood vessels, nerves, and nerve trajectories. To take full advantage of this resolution and realism, our collaborators at ZMT Zurich MedTech have empowered these models with state-of-the-art physics solvers (electromagnetic, thermal, acoustic, and computational fluid dynamics) and tissue models, thereby overcoming the challenges encountered during model manipulation in existing commercial platforms.
The ViP V2.0 models - Ella, Duke, Billie, and Thelonious - consist of simplified CAD files optimized for finite-element modeling in third-party commercial platforms such as ANSYS and CST. For the purpose of simplification, the original geometry of the approximately 300 organs and tissues of version 3.0 are combined into 22 high-resolution tissue groups.
The V3.x models are validated and the compatibility between versions has been assessed for EM simulations, i.e., the results of MR safety and MR implant safety evaluations are statistically identical for the ViP3.x and 1.x models. Note, however, that we do not warranty compatibility of the ViP V2.0 because they consist of simplified tissues.
The Virtual Population models V1-x and V4-x are fully compatible with the Poser tool available in the simulation platform Sim4Life. This tool allows postures to be modified for simulating various exposure configurations.
We remain dedicated to providing the community with new generations of increasingly more precise and more detailed 3D anatomical models and high-resolution tissue models along with a continually expanding and updated database of biological tissues properties. However, previous versions of our models remains available and are now unambiguously traceable.